
Prognostic value of Epstein-Barr virus biomarkers for nasopharyngeal carcinoma staging and post-treatment surveillance
Background
There is marked geographic variation in the incidence of nasopharyngeal carcinoma (NPC), with a lifetime risk as high as 1.8% in Guangdong, China (1). The Epstein-Barr virus (EBV)-associated variant of NPC comprises nearly all cases in endemic regions (2). This epidemiologic association between EBV and NPC was first observed decades ago, and since that time numerous EBV-based biomarkers have been developed to detect anti-EBV antibodies, circulating EBV nucleic acids, and circulating tumor cells (CTCs) (3-7). Although EBV is a nearly ubiquitous infection among adults worldwide, EBV-associated NPC has extreme regional and ethnic restriction that remains largely unexplained (8). Although human genome-wide association studies have identified human germline loci with modest attributable risk for NPC, select high-risk EBV variants appear to confer much more substantial risk in high-incidence populations (9,10).
Following primary infection, EBV undergoes lytic replication in the pharynx and establishes lifelong latent infection in B cells as an episome (11). EBV within latently-infected B cells can intermittently reactivate and replicate, shedding virions into the bloodstream that may reinfect epithelial cells. Importantly, EBV is not routinely detected in benign nasopharyngeal epithelium, suggesting that host or environmental factors must contribute to the persistent epithelial latent infection that precedes neoplasia. Clinically, the presence of EBV within NPCs can be demonstrated by fluorescence in situ hybridization of abundant non-coding RNAs (EBERs). It is this interaction between germline/environmental epithelial susceptibility and the oncogenic properties of EBV that is the current model of NPC pathogenesis (12).
Worldwide, NPC is the second leading cause of head and neck cancer mortality (13). Because NPC has a propensity for nodal metastasis, most unscreened patients present with locoregionally-advanced disease, for which the current standard of care is intensity-modulated concurrent chemoradiotherapy (CCRT) with or without induction (IC) or adjuvant chemotherapy (AC) (14-16). Although this approach achieves high rates of locoregional control, up to 30% of patients develop distant metastasis (14). Complex multidisciplinary care is required for treatment of these patients, and the aforementioned EBV-based biomarkers are increasingly recognized for their potential to guide risk-adapted treatment intensification or de-intensification. Similarly, many have proposed integrating these biomarkers into the traditional American Joint Committee on Cancer (AJCC) anatomic tumor, nodal, metastasis (TNM) staging system for improved prognostication, discrimination, and consistency (17,18).
In this review, we discuss the clinical role and prognostic performance of EBV-based biomarkers for pre-treatment staging and post-treatment surveillance. In particular, we synthesize the available evidence which suggests that biomarker-informed staging systems might improve upon anatomic staging, but highlight the challenges in inter-laboratory reproducibility inherent to diagnostic assays without international standardization. Thereafter, we review a breadth of evidence which supports that undetectable post-treatment EBV biomarkers are highly specific for long-term cure. Finally, we contextualize emerging biomarkers that may further improve prognostication.
Plasma EBV DNA PCR testing and harmonization
Although serologic EBV assays have been widely studied for early detection of preclinical NPC, these assays have had limited utility in pre-treatment prognostication and surveillance, in contrast to nucleic acid amplification tests (19). A quantitative real-time polymerase chain reaction (qPCR) to noninvasively detect cell-free EBV DNA in plasma was first described more than 20 years ago (4). This specific method amplifying the BamHI-W tandem-repeated fragment has been the cornerstone of biomarker-informed pre-treatment prognostication, post-treatment surveillance, and ongoing risk-adapted clinical trials. In contrast to commercial assays which amplify single-copy regions of the EBV genome encoding viral proteins (EBNA-1, LMP-1), the tandem-repeated nature of the BamHI-W sequence facilitates increased analytical and clinical sensitivity, although the number of copies per EBV genome may vary (20,21). After development of the BamHI-W qPCR assay by Lo and colleagues (4), Chan and colleagues further characterized the nature of EBV DNA in plasma and demonstrated that it remains in the supernatant and not the pellet after ultracentrifugation (22). With additional experiments that revealed the majority of DNA fragments in plasma were shorter than 181 nucleotides, it was deduced that EBV DNA in plasma is naked and not contained within intact virions. As such, EBV DNA in the plasma of patients with NPC does not suggest the presence of circulating virions but rather reflects cell-free DNA shed from infected neoplastic epithelium. Although whole blood may have certain advantages over plasma for detection of EBV DNA in the post-transplantation setting, the presence of latently-infected lymphocytes in whole blood would result in false positives in the NPC patient population, as cell-free EBV DNA is derived from epithelium (23-25).
While plasma and serum have been the primary matrices for population-level screening, nasopharyngeal PCR has also been investigated to triage screen-detected patients or detect local recurrences after radiotherapy (26,27). However, nasopharyngeal or salivary swabs alone are inadequate for surveillance or pre-treatment prognostication due to the propensity for NPC to metastasize to lymph nodes and distant organs. Accordingly, the presence of cell-free DNA in plasma facilitates monitoring of nearly all tissues in totality, which is in contrast to pharyngeal specimens. Many investigators have leveraged this to explore plasma EBV DNA as not only a qualitative biomarker for presence of malignancy but also a quantitative biomarker as a surrogate for disease burden before, during, and after radiotherapy (28). For example, Lv and colleagues described four prognostic NPC phenotypes based on plasma EBV DNA response during induction chemotherapy, and propose that these phenotypes be leveraged in treatment intensification or de-intensification protocols (29). As the burden of neoplastic cells decreases or increases in a given patient, there is a concomitant proportional change in cell-free EBV DNA that is detected in the quantitative PCR reaction.
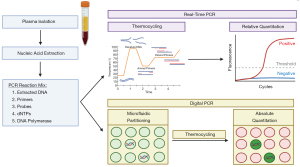
Notably, EBV PCR testing has yet to be standardized across institutions and myriad approaches to prognostication before, during, and after radiotherapy have been assessed. The critical importance of assay harmonization for biomarker-informed management has been highlighted in multi-institutional studies that demonstrate significant inter-laboratory quantitative variability (6). Typically, a log-transformed coefficient of variation below 20% within and between laboratories is desirable for quantitative PCR assays. Reproducibility can be improved with harmonization of external calibrators and reagents used in qPCR, and formal recommendations from a National Cancer Institute-convened workshop have been made to promote assay standardization (30). While post-transplantation EBV monitoring is routinely reported in WHO international units traceable to the NIBSC EBV international standard, this practice has not been adopted for NPC despite variability introduced by external calibrators (21,31). Although digital PCR may permit absolute quantitation without these external calibrators, the inter-laboratory performance of digital vs. real-time PCR remains unknown (Figure 1). With increasing multi-institutional evidence that a biomarker-informed staging system offers superior prognostication, assay standardization is of paramount importance.
Pre-treatment prognostication and biomarker-informed staging
The earliest studies quantifying EBV DNA in plasma noted differences in pre-treatment copy number among patients with early vs. advanced anatomic stage and presence or absence of post-treatment relapse (4,32). These investigators recognized that EBV DNA copy number in plasma correlated with tumor burden and was also prognostic for long-term outcome. Since that time, numerous studies have assessed the prognostic value of plasma EBV DNA copy number adjusted for anatomic stage, and several large studies have proposed biomarker-informed staging systems based on partitioning analyses.
Before assessing the prognostic value of pre-treatment EBV DNA, it is important to consider differences among studies in inclusion criteria, staging systems, imaging modalities, treatment, and plasma EBV DNA quantitation (Figure 2) (3,18,32-37). While all studies amplify the BamHI-W fragment, variable volume of extracted plasma (200–800 µL), elution volumes (50–100 µL), eluate volume in the qPCR reaction (2–10 µL), and reagents/calibrators could contribute to imprecision or systematic differences in quantitative accuracy that yield differing prognostic cutoffs (500–40,000 copies/mL). With the exception of the earliest series, most studies propose that 1,500–4,000 cp/mL is associated with an increased risk of relapse, with generally similar hazard ratios. Importantly, the optimal cutoff may vary among these studies due to inherent differences in stage distributions and treatment paradigms across institutions and time periods. The pooled results of these select studies across institutions, staging systems, and treatment paradigms indicate a relatively consistent progression-free survival (PFS) hazard ratio (Figure 2). These findings are similar to a systematic review and meta-analysis including studies published from 2001–2014 (37).

Since 2015, there have been several large studies conducted with the specific objective of improving upon the AJCC anatomic staging system via integration of pre-treatment plasma EBV DNA. In 2015, Tang et al. published a study including more than 6,300 patients treated at Sun Yat-sen University Cancer Center (SYSUCC) spanning a training cohort (n=3,113), internal validation cohort (n=1,556), and prospective validation cohort (n=1,668) (36). Patients were treated according to the existing evidence at that time, with either radiotherapy alone (AJCC 7 stage I, select stage II), concurrent chemoradiotherapy alone (select stage II), or chemoradiotherapy with induction or AC (stage III-IV). Based on prior evidence, a cutoff of 4,000 cp/mL was assessed, which was independently prognostic for disease-free survival, distant-metastasis-free survival, and overall survival (OS) after adjusting for other prognosticators in both the training and validation datasets. In addition, high-sensitivity CRP (hs-CRP) was also independently prognostic in this study. Based on these findings, the authors proposed and validated a new risk stratification wherein patients with early (stage I-II) or advanced (stage III-IV) disease were categorized by low vs. high hs-CRP and/or EBV DNA. An open question is whether hs-CRP (HR 1.82) has sufficient additive prognostic value to EBV DNA (HR 2.99) for widespread adoption, as it is not routinely collected during workup.
In 2019, Lee and colleagues published results from an institutional prospective cohort study with the intention of improving upon the AJCC 8 staging system (18,38). In this cohort of 518 patients with nonmetastatic NPC treated at The University of Hong Kong with definitive intensity-modulated radiotherapy (IMRT), recursive partitioning analysis (RPA) identified 536 cp/mL as the optimal cutoff. Out of simplicity, the authors reasonably selected 500 cp/mL for further evaluation and internal validated this cutoff in bootstrap analysis. As shown in Table 1, the final RPA created five stage groups [AJCC 8 T1-4N0-2 with EBV <500 cp/mL (I), T1-4N0-2 with EBV ≥500 cp/mL (II), T1-2N3 (III), T3-4N3 (IVA), and T1-4N0-3M1 (IVB)].
Table 1
| AJCC 8 T stage | AJCC 8 N stage | AJCC 8 M stage | Plasma EBV DNA (copies/mL) | AJCC 8 TNM | Lee et al. 2019 | Guo et al. 2019 |
|---|---|---|---|---|---|---|
| T1 | N0 | M0 | Any | I | – | – |
| T1 | N1 | M0 | Any | II | – | – |
| T2 | N0–1 | M0 | Any | II | – | – |
| T1-2 | N2 | M0 | Any | III | – | – |
| T3 | N0–2 | M0 | Any | III | – | – |
| T4 | Any N | M0 | Any | IVA | – | – |
| Any T | N3 | M0 | Any | IVA | – | – |
| Any T | Any N | M1 | Any | IVB | – | – |
| Any T | N0–2 | M0 | <500 | – | I | – |
| Any T | N0–2 | M0 | ≥500 | – | II | – |
| T1-2 | N3 | M0 | Any | – | III | – |
| T3-4 | N3 | M0 | Any | – | IVA | – |
| Any T | Any N | M1 | Any | – | IVB | – |
| T1 | N0 | M0 | Any | – | – | I |
| T2-3 | N0 | M0 | ≤2,000 | – | – | IIA |
| T1-3 | N1 | M0 | ≤2,000 | – | – | IIA |
| T2-3 | N0 | M0 | >2,000 | – | – | IIB |
| T1-3 | N1 | M0 | >2,000 | – | – | IIB |
| T1-3 | N2 | M0 | ≤2,000 | – | – | IIB |
| T1-3 | N2 | M0 | >2,000 | III | ||
| T4 | N0–2 | M0 | Any | – | – | III |
| Any T | N3 | M0 | Any | – | – | IVA |
| Any T | Any N | M1 | Any | – | – | IVB |
TNM, tumor, node, metastasis; AJCC, American Joint Committee on Cancer; EBV, Epstein-Barr virus.
In a rigorous comparison of the biomarker-informed RPA against the AJCC 8 system, the RPA offered superior hazard consistency, hazard discrimination, explanation of variance, and overall performance for PFS, OS, and cancer-specific survival (CSS). Performance of the RPA was also similar or superior to a biomarker-informed multivariable Cox model, which would be one alternative to partitioning analysis. A clinically-relevant consequence of this RPA was the observation that RPA stage I patients using their proposed staging system (AJCC 8 T1-4N0-2 with EBV <500 cp/mL) appeared to derive marginal benefit from the addition of chemotherapy to radiotherapy, highlighting opportunities for risk-adapted clinical trials.
Finally, Guo and colleagues reported results from a similarly-designed study which included 979 patients in a retrospective training cohort and 550 patients in a prospective validation cohort treated at SYSUCC (Table 1) (17). An EBV DNA cutoff of 2,000 cp/mL was selected based on prior institutional experience (39). Following RPA, five revised biomarker-informed stages were proposed: T1N0M0 (I), T1-3N0-1M0 and EBV DNA ≤2,000 (IIA), T1-3N0-1M0 and EBV DNA >2,000 or T1-3N2M0 and EBV DNA ≤2,000 (IIB), T1-3N2M0 and EBV DNA >2,000 or T4N0-2M0 (III), T1-4N3M0 (IVA), and T1-4N0-3M1 (IVB). As similarly reported by Lee et al., the biomarker-informed system offered improved hazard consistency, hazard discrimination, explanation of variance, and overall performance. The authors propose several specific recommendations for a future biomarker-informed system. With the routine use of IMRT, the authors observed minimal independent discrimination between T2 and T3 tumors, both of which had high rates of locoregional control. Furthermore, patients with bilateral (N2) nodal disease and low EBV DNA (<2,000 cp/mL) did not exhibit higher rates of distant metastasis relative to patients with N0-1 disease. These findings suggest that the T4 and N3 categories are the most pertinent anatomic staging factors followed by EBV DNA, whereas the prognostic value of the remaining anatomic factors (T1-3N0-2) are relatively minor and conditional on EBV DNA.
Broadly, these select studies in combination with other institutional experiences suggest that a pre-treatment plasma EBV DNA cutoff ranging from 500–4,000 cp/mL is an independent prognosticator for PFS despite variable inclusion criteria, staging systems/modalities, treatment paradigms, and qPCR assays. The aforementioned biomarker-informed staging systems developed via partitioning analysis have the potential to identify groups of patients with similar prognosis despite differing anatomic stage. For example, a recently-presented phase II trial randomized patients with AJCC 7 stage III-IVB NPC and EBV DNA <4,000 cp/mL to either two or three cycles of 100 mg/m2 cisplatin with concurrent radiotherapy (40). With a median follow-up of nearly three years, two cycles of cisplatin was non-inferior to the standard three cycles and had reduced toxicity. Additional clinical trials are awaited to validate high- and low-risk biomarker groups through selective treatment intensification and de-intensification.
Post-treatment prognostication and surveillance
Circulating cancer antigen biomarkers have long been used for pre-treatment prognostication, response assessment, and post-treatment surveillance in prostate specific antigen (PSA), ovarian (CA-125), and gastrointestinal [carcinoembryonic antigen (CEA), CA 19-9] cancers. Similarly, the phenomenon of plasma EBV DNA clearance after radiotherapy has long been recognized as a favorable prognosticator (32). Although most patients do achieve EBV DNA clearance, a subset experience biomarker persistence/recurrence which generally precedes clinical relapse by weeks to months. The sensitivity and specificity of post-treatment EBV DNA for relapse has therefore motivated cooperative-group clinical trials and staging systems for post-treatment prognostication.
Although many institutions practice routine EBV DNA surveillance, it is important to contextualize this biomarker alongside standard imaging-based response assessment. In conjunction with physical examination and nasoendoscopy, NCCN guidelines recommend post-treatment FDG-PET/CT for confirmation of clinical complete response, with EBV DNA surveillance as a category 2B recommendation (41). In a systematic review and meta-analysis of studies assessing the diagnostic accuracy of FDG-PET, CT, and MRI for diagnosis of residual/recurrent NPC, the sensitivity/specificity of PET (95%/90%) exceeded both CT (76%/59%) and MRI (78%/76%) (42). While EBV DNA is a relatively sensitive and specific biomarker for recurrence, select series have suggested low sensitivity (52%) for local recurrence, highlighting the importance of confirming clinical complete response and continuing clinic-based surveillance (43).
Multiple studies have identified post-treatment EBV DNA clearance (0 cp/mL) as most prognostic for PFS (Figure 3). These publications also differed in study design, inclusion criteria, staging systems/modalities, treatment paradigms, timing of plasma collection, and EBV PCR assays. EBV DNA clearance generally has higher specificity for relapse than pre-treatment EBV DNA, and therefore is more prognostic but perhaps less actionable than pre-treatment EBV DNA. In a meta-analysis of six studies published between 2002 and 2014, the pooled hazard ratios for OS and PFS were 4.26 and 5.21 among patients with detectable post-treatment EBV DNA, relative to 2.81 and 2.74 for high vs. low pre-treatment EBV DNA. Ongoing efforts seek to integrate longitudinal biomarker response before, during, and after chemoradiotherapy for adaptive treatment (29).

A critical consideration when evaluating these studies is the timing of post-treatment plasma collection for EBV DNA PCR. Similar to the timing of post-treatment imaging for response assessment in head and neck carcinomas, the timing of response assessment could impact sensitivity and specificity, with a greater proportion of false positives if early response assessment is conducted (44). To date, existing studies have collected plasma for EBV DNA PCR as early as seven days and as late as four months after radiotherapy (29,33,45). Furthermore, many studies report collection of plasma “within three months” after radiotherapy, which may introduce heterogeneity in biomarker performance (35,46,47). The prospective randomized HKNPCSG-0502 trial and an early prospective observational study mandated plasma collection 6–8 weeks after radiotherapy, whereas the ongoing NRG-HN001 trial mandates collection within one week after radiotherapy (3,48,49).
Among the earliest publications identifying post-treatment EBV DNA clearance as prognostic was a prospective observational series reported by Chan et al. (3) The authors recruited 170 patients with AJCC 5th edition stage II-IVB NPC treated with concurrent chemoradiotherapy. With a median follow-up of 27 months at the time of publication, post-treatment EBV DNA >500 cp/mL was highly prognostic for PFS (HR 11.9). The sensitivity and specificity for relapse at this cutoff were 67% and 94%, respectively. This study, among others, hypothesized that patients with a favorable biomarker response might be spared AC given low rates of relapse in this group.
The HKNPCSG-0502 randomized trial is the highest level of evidence thus far for the prognostic and predictive role of post-treatment EBV DNA (49). This trial enrolled 789 patients with AJCC 6th edition stage IIB-IVB NPC across all six oncology centers in Hong Kong. The primary objective of the study was to determine if adjuvant gemcitabine/cisplatin, which has efficacy in the metastatic setting, would improve relapse-free survival among patients without EBV DNA clearance (16). The study was designed with separate prospective observational and randomized arms, wherein patients with undetectable EBV DNA 6–8 weeks after radiotherapy were observed and patients with persistently-detectable EBV DNA were randomly assigned to observation or six cycles of adjuvant gemcitabine/cisplatin. Patients were permitted to receive radiotherapy alone (19%), concurrent chemoradiotherapy (81%), and/or neoadjuvant chemotherapy (25%).
After radiotherapy, 573 (73%) patients had no detectable EBV DNA and were observed. This cohort had excellent 5-year OS (87%) which was not significantly different than patients with 1–49 cp/mL (83%). In contrast, patients with 50–499 cp/mL or ≥500 cp/mL had significant worse 5-year survival (51% and 27%, respectively).
Among the remaining 216 patients (27%) with persistently-detectable EBV DNA, 112 were excluded for randomization (patient refusal, residual disease, distant metastasis, or renal/hematologic function) and 104 (13%) were randomized. In the 52 patients randomized to adjuvant gemcitabine/cisplatin, 50% completed all six cycles and 65% completed at least four cycles. Tolerance to AC was lower than in the metastatic setting (83% four cycles, 58% six cycles), likely due to radiotherapy-associated acute toxicities. With a median follow-up of 6.6 years, there was no improvement in relapse-free survival between the randomized arms. The authors postulated that several factors might have contributed to the trial results, including AC compliance, duration between completing radiotherapy and initiation of chemotherapy (median 91 days), exclusion of patients at higher risk for relapse in the prerandomization evaluation, and inability to eradicate platinum-resident clones with gemcitabine/platinum.
The ongoing NRG-HN001 trial randomizes a similar population of patients with persistently-detectable EBV DNA to non-cross-resident paclitaxel/gemcitabine at an earlier time point (28 days after radiotherapy), which will add further clarity to the predictive vs. prognostic significance of post-treatment EBV DNA. Similar to HKNPCSG-0502, The National Health Research Institutes of Taiwan are also enrolling patients with detectable post-treatment EBV DNA on a randomized phase III trial of observation vs. adjuvant MEP chemotherapy followed by oral Tegafur-uracil (50). Finally, SYSUCC is enrolling a similar population of patients on a II randomized trial of observation vs. oral apatinib (51).
Across the selected studies in Figure 3, the pooled sensitivity and specificity for relapse of post-treatment EBV DNA is 72% and 81%, highlighting opportunities for further improvements to biomarker-informed surveillance programs. Chen et al. reported results from a large series of 1,984 patients treated at SYSUCC, among which 767 (39%) had detectable EBV DNA after radiotherapy (47). Importantly, the sensitivity for detection of local recurrence (69%) was lower than for regional (80%) or distant (91%) recurrence, concordant with Leung et al. (43). Biomarker relapse preceded clinical relapse by a median of 2.3 months, and 82% of patients who had detectable EBV DNA but did not develop relapse cleared EBV DNA during long-term monitoring. These findings have implications for surveillance programs, as a significant proportion of patients with local recurrence can be successfully salvaged, highlighting the importance of early endoscopic and/or imaging-based detection (52).
Novel biomarkers for improved prognostication
Plasma EBV BamHI-W DNA remains the most widely utilized biomarker for EBV-associated NPC before, during, and after definitive therapy. However, given the opportunity to improve upon its sensitivity and specificity for relapse, many groups have explored novel or complementary biomarkers that include microRNAs (miRNA), CTCs, and serologic assays.
The EBV genome encodes dozens of BART and BHRF1 miRNAs related to viral gene expression and post-transcriptional modification. In 2012, Liu and colleagues published findings from a miRNA expression analysis of 312 paraffin-embedded NPC tissue specimens collected at SYSUCC, which were compared against 18 specimens of benign nasopharyngitis (53). Forty-one miRNAs were differentially expressed in NPC, and a risk score comprising five miRNAs (miR-93, miR-142-3p, miR-29c, miR-26a, miR-30e) was found to be independently prognostic for disease-free, distant metastasis-free, and OS after adjusting for anatomic stage and other clinical prognosticators. It remains to be determined whether this miRNA signature has prognostic value in addition to plasma EBV DNA, and whether this panel of miRNAs can be detected in plasma or standardized across laboratories.
Several groups have also studied circulating EBV miRNAs and their diagnostic and prognostic performance against BamHI-W EBV DNA. Zhang et al. profiled EBV miRNA expression in EBV latently-infected cell lines, an NPC-derived cell line, and an artificially-infected NP epithelium cell line (54). After identifying differentially-expressed miRNAs, a case-control series demonstrated that a combination of miR-BART7-3p and miR-BART13-3p had 90% accuracy for distinguishing 89 NPC patients from 28 healthy controls. The author subsequently validated these findings in a larger study of 465 NPC nonmetastatic patients and 243 healthy controls (55). In this study, the sensitivities and specificities of miR-BART7-3p and miR-BART13-3p for NPC were each 96–98%, which was marginally improved over EBV DNA (94% sensitive, 91% specific). The combination of these three nucleic acids yielded better diagnostic accuracy than any single biomarker, with an AUC of 0.997 for NPC. Similar to EBV DNA, most NPC patients achieved miR-BART7-3p clearance (82%) and miR-BART13-3p clearance (45%) after radiotherapy. Persistently-detectable EBV DNA and miR-BART7-3p, but not miR-BART13-3p, were independently prognostic for poorer distant metastasis-free survival relative to patients achieving biomarker clearance. Moreover, the combination of EBV DNA and miR-BART7-3p had greater prognostic value than either nucleic acid alone, highlighting opportunities for further risk stratification in clinical trials.
In contrast to nucleic acid amplification techniques, CTCs have potential utility as both prognostic and functional biomarkers. Few studies have characterized CTCs in NPC, but preliminary evidence suggests they may be detectable in most patients. Zhang and colleagues enumerated NPC-associated CTCs using subtraction-enrichment fluorescence in situ hybridization, and observed that 92% of patients had identifiable CTCs (56). Importantly, CTCs were defined only by the presence of nucleated cells positive for EpCAM and/or chromosome 8 aneuploidy without CD45, which may not be specific to NPC and might be observed in non-NPC controls or other epithelial malignancies. The investigators observed differential CTC dynamics among patients with or without response to induction chemotherapy, and also reported that aneuploidy appeared to functionally correlate with response to chemotherapy. Because the equipment and technical expertise for CTC detection are not widely available, further studies will be required to determine the role of this biomarker in pre-treatment prognostication and post-treatment surveillance.
Due to heterogeneity in study design, laboratory methodology, inclusion criteria, and statistical analysis, it remains difficult to discern the relative performance of these novel biomarkers in the context of longstanding biomarkers such as BamHI-W DNA, EBNA-1 DNA, and anti-VCA/EA IgA. For this reason, Tan and colleagues conducted a systematic comparison of ten distinct EBV DNA, miRNA, and serologic biomarkers characterized from the same specimens (57). In a large cohort of 251 healthy controls and 232 patients with NPC, the 76-nucleotide BamHI-W amplicon had the highest accuracy (96%), which was greater than the 121-nucleotide BamHI-W amplicon (90%), EBNA-1 (94%), miRNA-BART7-3p (86%), and anti-VCA/EA IgA (57–65%), among others. Because BamHI-W may be detected in healthy controls, partitioning analysis suggested that a combination of BamHI-W and either anti-VCA IgA or anti-EA IgG may slightly improve specificity. This approach has previously been studied in the context of early NPC detection during population-level screening (58).
Conclusions and future directions
The relationship between EBV infection and endemic NPC has facilitated more than 40 years of biomarker-inspired translational research. Serologic and nucleic acid EBV biomarkers span the spectrum of this disease from population-level early detection, pre-treatment prognostication, response-adapted therapy, and long-term surveillance. Plasma EBV DNA remains the cornerstone of biomarker prognostication and surveillance, and there is increasing high-quality evidence that it merits inclusion in future staging systems. However, inter-laboratory reproducibility remains largely unaddressed, while imperfect sensitivity and specificity highlight opportunities for novel complementary biomarkers to further risk stratify patients. Ongoing and future clinical trials will determine whether biomarker-adapted management will be the standard of care.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Maria Li Lung, Lawrence S. Young) for the series “NPC Biomarkers” published in Annals of Nasopharynx Cancer. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://anpc.amegroups.com/article/view/10.21037/anpc-21-5/coif). The series “NPC Biomarkers” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Colombet M, Mery L, et al. Cancer Incidence in Five Continents, Vol. XI [Internet]. Lyon: International Agency for Research on Cancer; 2017 [cited 2019 Jan 20]. Available online: http://ci5.iarc.fr
- El-Naggar A, Chan J, Grandis J, et al. WHO Classification of Head and Neck Tumours [Internet]. [cited 2019 Jan 20]. Available online: http://publications.iarc.fr/Book-And-Report-Series/Who-Iarc-Classification-Of-Tumours/Who-Classification-Of-Head-And-Neck-Tumours-2017
- Chan AT, Lo YM, Zee B, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst 2002;94:1614-9. [Crossref] [PubMed]
- Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 1999;59:1188-91. [PubMed]
- Chan KH, Gu YL, Ng F, et al. EBV specific antibody-based and DNA-based assays in serologic diagnosis of nasopharyngeal carcinoma. Int J Cancer 2003;105:706-9. [Crossref] [PubMed]
- Le QT, Zhang Q, Cao H, et al. An international collaboration to harmonize the quantitative plasma Epstein-Barr virus DNA assay for future biomarker-guided trials in nasopharyngeal carcinoma. Clin Cancer Res 2013;19:2208-15. [Crossref] [PubMed]
- Li F, Liu J, Song D, et al. Circulating tumor cells in the blood of poorly differentiated nasal squamous cell carcinoma patients: correlation with treatment response. Acta Otolaryngol 2016;136:1164-7. [Crossref] [PubMed]
- Smatti MK, Al-Sadeq DW, Ali NH, et al. Epstein-Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population: An Update. Front Oncol 2018;8:211. [Crossref] [PubMed]
- Xu M, Yao Y, Chen H, et al. Genome sequencing analysis identifies Epstein-Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat Genet 2019;51:1131-6. [Crossref] [PubMed]
- Su WH, Hildesheim A, Chang YS. Human leukocyte antigens and epstein-barr virus-associated nasopharyngeal carcinoma: old associations offer new clues into the role of immunity in infection-associated cancers. Front Oncol 2013;3:299. [Crossref] [PubMed]
- Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 2016;16:789-802. [Crossref] [PubMed]
- Wong KCW, Hui EP, Lo KW, et al. Nasopharyngeal carcinoma: an evolving paradigm. Nat Rev Clin Oncol 2021; Epub ahead of print. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Pan JJ, Ng WT, Zong JF, et al. Prognostic nomogram for refining the prognostication of the proposed 8th edition of the AJCC/UICC staging system for nasopharyngeal cancer in the era of intensity-modulated radiotherapy. Cancer 2016;122:3307-15.
- Yang Q, Cao SM, Guo L, et al. Induction chemotherapy followed by concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: long-term results of a phase III multicentre randomised controlled trial. Eur J Cancer 2019;119:87-96. [Crossref] [PubMed]
- Zhang Y, Chen L, Hu GQ, et al. Gemcitabine and Cisplatin Induction Chemotherapy in Nasopharyngeal Carcinoma. N Engl J Med 2019;381:1124-35. [Crossref] [PubMed]
- Guo R, Tang LL, Mao YP, et al. Proposed modifications and incorporation of plasma Epstein-Barr virus DNA improve the TNM staging system for Epstein-Barr virus-related nasopharyngeal carcinoma. Cancer 2019;125:79-89. [Crossref] [PubMed]
- Lee VH, Kwong DL, Leung TW, et al. The addition of pretreatment plasma Epstein-Barr virus DNA into the eighth edition of nasopharyngeal cancer TNM stage classification. Int J Cancer 2019;144:1713-22.
- Ji MF, Sheng W, Cheng WM, et al. Incidence and mortality of nasopharyngeal carcinoma: interim analysis of a cluster randomized controlled screening trial (PRO-NPC-001) in southern China. Ann Oncol 2019;30:1630-7. [Crossref] [PubMed]
- Sanosyan A, Fayd'herbe de Maudave A, Bollore K, et al. The impact of targeting repetitive BamHI-W sequences on the sensitivity and precision of EBV DNA quantification. PLoS One 2017;12:e0183856. [Crossref] [PubMed]
- Abeynayake J, Johnson R, Libiran P, et al. Commutability of the Epstein-Barr virus WHO international standard across two quantitative PCR methods. J Clin Microbiol 2014;52:3802-4. [Crossref] [PubMed]
- Chan KC, Zhang J, Chan AT, et al. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res 2003;63:2028-32. [PubMed]
- Fafi-Kremer S, Brengel-Pesce K, Barguès G, et al. Assessment of automated DNA extraction coupled with real-time PCR for measuring Epstein-Barr virus load in whole blood, peripheral mononuclear cells and plasma. J Clin Virol 2004;30:157-64. [Crossref] [PubMed]
- Stevens SJ, Pronk I, Middeldorp JM. Toward standardization of Epstein-Barr virus DNA load monitoring: unfractionated whole blood as preferred clinical specimen. J Clin Microbiol 2001;39:1211-6. [Crossref] [PubMed]
- Wadowsky RM, Laus S, Green M, et al. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J Clin Microbiol 2003;41:5245-9. [Crossref] [PubMed]
- Chen Y, Zhao W, Lin L, et al. Nasopharyngeal Epstein-Barr Virus Load: An Efficient Supplementary Method for Population-Based Nasopharyngeal Carcinoma Screening. PLoS One 2015;10:e0132669. [Crossref] [PubMed]
- Adham M, Greijer AE, Verkuijlen SA, et al. Epstein-Barr virus DNA load in nasopharyngeal brushings and whole blood in nasopharyngeal carcinoma patients before and after treatment. Clin Cancer Res 2013;19:2175-86. [Crossref] [PubMed]
- Huang CL, Sun ZQ, Guo R, et al. Plasma Epstein-Barr Virus DNA Load After Induction Chemotherapy Predicts Outcome in Locoregionally Advanced Nasopharyngeal Carcinoma. Int J Radiat Oncol Biol Phys 2019;104:355-61. [Crossref] [PubMed]
- Lv J, Chen Y, Zhou G, et al. Liquid biopsy tracking during sequential chemo-radiotherapy identifies distinct prognostic phenotypes in nasopharyngeal carcinoma. Nat Commun 2019;10:3941. [Crossref] [PubMed]
- Kim KY, Le QT, Yom SS, et al. Current State of PCR-Based Epstein-Barr Virus DNA Testing for Nasopharyngeal Cancer. J Natl Cancer Inst 2017;109:djx007. [Crossref] [PubMed]
- Fryer JF, Heath A, Wilkinson DE, et al. Collaborative study to evaluate the proposed 1st WHO international standards for Epstein-Barr Virus (EBV) for Nucleic Acid Amplification Technology (NAT)-Based Assays. Geneva: World Health Organization; 2011.
- Lo YM, Chan AT, Chan LY, et al. Molecular prognostication of nasopharyngeal carcinoma by quantitative analysis of circulating Epstein-Barr virus DNA. Cancer Res 2000;60:6878-81. [PubMed]
- Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med 2004;350:2461-70. [Crossref] [PubMed]
- Leung SF, Zee B, Ma BB, et al. Plasma Epstein-Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J Clin Oncol 2006;24:5414-8. [Crossref] [PubMed]
- Leung SF, Chan KC, Ma BB, et al. Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann Oncol 2014;25:1204-8. [Crossref] [PubMed]
- Tang LQ, Li CF, Chen QY, et al. High-sensitivity C-reactive protein complements plasma Epstein-Barr virus deoxyribonucleic acid prognostication in nasopharyngeal carcinoma: a large-scale retrospective and prospective cohort study. Int J Radiat Oncol Biol Phys 2015;91:325-36. [Crossref] [PubMed]
- Zhang W, Chen Y, Chen L, et al. The clinical utility of plasma Epstein-Barr virus DNA assays in nasopharyngeal carcinoma: the dawn of a new era?: a systematic review and meta-analysis of 7836 cases. Medicine (Baltimore) 2015;94:e845. [Crossref] [PubMed]
- Pan JJ, Ng WT, Zong JF, et al. Proposal for the 8th edition of the AJCC/UICC staging system for nasopharyngeal cancer in the era of intensity-modulated radiotherapy. Cancer 2016;122:546-58.
- Peng H, Guo R, Chen L, et al. Prognostic Impact of Plasma Epstein-Barr Virus DNA in Patients with Nasopharyngeal Carcinoma Treated using Intensity-Modulated Radiation Therapy. Sci Rep 2016;6:22000. [Crossref] [PubMed]
- Mai HQ, Li XY, Mo HY, et al. De-intensified chemoradiotherapy for locoregionally advanced nasopharyngeal carcinoma based on plasma EBV DNA: A phase 2 randomized noninferiority trial. [cited 2021 May 23]. Available online: https://meetinglibrary.asco.org/record/196656/abstract
- NCCN Clinical Practice Guidelines in Oncology. Head and Neck Cancers. Version 1.2021. [Internet]. National Comprehensive Cancer Network (NCCN); 2020 [cited 2021 Mar 18]. Available online: https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf
- Liu T, Xu W, Yan WL, et al. FDG-PET, CT, MRI for diagnosis of local residual or recurrent nasopharyngeal carcinoma, which one is the best? A systematic review. Radiother Oncol 2007;85:327-35. [Crossref] [PubMed]
- Leung SF, Lo YM, Chan AT, et al. Disparity of sensitivities in detection of radiation-naïve and postirradiation recurrent nasopharyngeal carcinoma of the undifferentiated type by quantitative analysis of circulating Epstein-Barr virus DNA1,2. Clin Cancer Res 2003;9:3431-4. [PubMed]
- Cheung PK, Chin RY, Eslick GD. Detecting Residual/Recurrent Head Neck Squamous Cell Carcinomas Using PET or PET/CT: Systematic Review and Meta-analysis. Otolaryngol Head Neck Surg 2016;154:421-32. [Crossref] [PubMed]
- Hui EP, Li WF, Ma BB, et al. Integrating postradiotherapy plasma Epstein-Barr virus DNA and TNM stage for risk stratification of nasopharyngeal carcinoma to adjuvant therapy. Ann Oncol 2020;31:769-79. [Crossref] [PubMed]
- Le QT, Jones CD, Yau TK, et al. A comparison study of different PCR assays in measuring circulating plasma epstein-barr virus DNA levels in patients with nasopharyngeal carcinoma. Clin Cancer Res 2005;11:5700-7. [Crossref] [PubMed]
- Chen FP, Huang XD, Lv JW, et al. Prognostic potential of liquid biopsy tracking in the posttreatment surveillance of patients with nonmetastatic nasopharyngeal carcinoma. Cancer 2020;126:2163-73. [Crossref] [PubMed]
- NRG Oncology. Randomized Phase II and Phase III Studies of Individualized Treatment for Nasopharyngeal Carcinoma Based on Biomarker Epstein Barr Virus (EBV) Deoxyribonucleic Acid (DNA) [Internet]. clinicaltrials.gov; 2021 Feb [cited 2021 May 6]. Report No.: NCT02135042. Available online: https://clinicaltrials.gov/ct2/show/NCT02135042
- Chan ATC, Hui EP, Ngan RKC, et al. Analysis of Plasma Epstein-Barr Virus DNA in Nasopharyngeal Cancer After Chemoradiation to Identify High-Risk Patients for Adjuvant Chemotherapy: A Randomized Controlled Trial. J Clin Oncol 2018; Epub ahead of print. [Crossref] [PubMed]
- National Health Research Institutes, Taiwan. Phase III Randomized Trial of Immediate Adjuvant Chemotherapy or Delayed Salvage Chemotherapy in Nasopharyngeal Carcinoma Patients With Post-radiation Detectable Plasma EBV DNA [Internet]. clinicaltrials.gov; 2015 Feb [cited 2021 May 6]. Report No.: NCT02363400. Available online: https://clinicaltrials.gov/ct2/show/NCT02363400
- Ma J. ADjuVant Apatinib in Nasopharyngeal Carcinoma Patients With Residual Epstein-Barr Virus (EBV) DNA Following Radiotherapy With or Without Chemotherapy [Internet]. clinicaltrials.gov; 2016 Oct [cited 2021 May 6]. Report No.: NCT02874651. Available online: https://clinicaltrials.gov/ct2/show/NCT02874651
- Liu YP, Wen YH, Tang J, et al. Endoscopic surgery compared with intensity-modulated radiotherapy in resectable locally recurrent nasopharyngeal carcinoma: a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol 2021;22:381-90. [Crossref] [PubMed]
- Liu N, Chen NY, Cui RX, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol 2012;13:633-41. [Crossref] [PubMed]
- Zhang G, Zong J, Lin S, et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer 2015;136:E301-12. [Crossref] [PubMed]
- Lu T, Guo Q, Lin K, et al. Circulating Epstein-Barr virus microRNAs BART7-3p and BART13-3p as novel biomarkers in nasopharyngeal carcinoma. Cancer Sci 2020;111:1711-23. [Crossref] [PubMed]
- Zhang J, Shi H, Jiang T, et al. Circulating tumor cells with karyotyping as a novel biomarker for diagnosis and treatment of nasopharyngeal carcinoma. BMC Cancer 2018;18:1133. [Crossref] [PubMed]
- Tan LP, Tan GW, Sivanesan VM, et al. Systematic comparison of plasma EBV DNA, anti-EBV antibodies and miRNA levels for early detection and prognosis of nasopharyngeal carcinoma. Int J Cancer 2020;146:2336-47. [Crossref] [PubMed]
- Ji MF, Huang QH, Yu X, et al. Evaluation of plasma Epstein-Barr virus DNA load to distinguish nasopharyngeal carcinoma patients from healthy high-risk populations in Southern China. Cancer 2014;120:1353-60. [Crossref] [PubMed]
Cite this article as: Miller JA, Pinsky BA, Le QT. Prognostic value of Epstein-Barr virus biomarkers for nasopharyngeal carcinoma staging and post-treatment surveillance. Ann Nasopharynx Cancer 2022;6:2.


