
Clinical utility of circulating tumor cells for real-time serial monitoring of nasopharyngeal carcinoma patients
Introduction
Nasopharyngeal carcinoma (NPC) is an endemic disease known as the “Guangdong” cancer and is prevalent in Southeast Asia, Middle East, and North Africa (1). In Hong Kong, the age-specific incidence rate for NPC in 2018 was ~30–35 per 100,000 for male patients aged 40–75 (2). In Hong Kong, NPC is amongst the top cancers in young males aged 20–44 (2). The distinctive geographical and ethnic variation of NPC incidence worldwide, together with characteristic features of ubiquitous latent Epstein-Barr virus (EBV) infection and the heavy lymphocyte infiltration distinguish NPC from other head and neck squamous cell carcinoma (HNSCC) (1). Important etiologic factors for NPC include latent EBV infection, host genetics and environmental factors. Since NPC is asymptomatic in early stages, about 80% of NPC patients are diagnosed at a late stage with locoregionally advanced metastatic disease to the cervical lymph nodes or distant organs. NPC patients have a higher tendency for distant metastasis among different types of head and neck cancers (3). Synchronous metastasis is detected in 4–10% NPC patients at presentation (4). Among locally advanced NPC patients treated by radical chemo-radiotherapy (CRT), 20–30% recur with metachronous metastasis within 3 years (5). These metastatic NPC (mNPC) patients treated by palliative chemotherapy (CT) have poor prognosis with median overall survival (OS) of 12–15 months (6). Despite efforts in genomic studies from ours and several other groups, which suggested the NFκB signaling pathway may be the driver in NPC carcinogenesis, the underlying molecular pathogenesis for mNPC is still poorly understood (7-12). A screening study conducted in Hong Kong utilizing the quantitative-polymerase chain reaction (Q-PCR) analysis of plasma EBV cell-free DNA (cfDNA) demonstrated the useful application for screening of asymptomatic NPC and the detection of NPC patients at an earlier stage for better outcome (13). Therefore, there are unmet needs for biomarkers for earlier detection of disease, stratification of patients for higher risk of relapse, real-time disease monitoring of minimal residual disease (MRD), prediction of treatment outcome, and surveillance of relapse. The identification of biomarkers from liquid biopsies which may be taken multiple times and real-time during treatment are expected to improve the survival of mNPC patients.
Circulating tumor cells (CTCs)
Although CTCs are regarded as the main seeds of distant metastases, the intrinsic accumulation of multiple genetic and epigenetic alterations is not sufficient to drive metastasis. The metastasis driving force requires the long-term continuous interaction with the exogenous stimuli produced by various tumor-associated stromal cells in the tumor microenvironment (TME) during the multi-step metastatic journey. The pro-tumorigenic inflammatory milieu of cytokines, chemokines, growth factors and proteases released by various tumor-associated stromal cells (tumor-associated endothelial cells, tumor-associated immune cells, and tumor-associated fibroblasts) foster a TME to induce epithelial tumor cells to undergo partial epithelial-mesenchymal transition (EMT) and dedifferentiation, giving rise to motile malignant carcinomas that have potential to breach the baseline membrane (14-16). In multicellular organisms, EMT is an important fundamental developmental program governing morphogenesis and granting motility and invasiveness properties to the polarized epithelial cells during embryonic development (17). EMT provides the basis for the dissociation of single carcinoma cells from the primary site (17). Most CTCs face strong selective forces after escape into the harsh conditions in the blood circulation from the primary or metastatic lesions; usually these cells die after few hours due to fluid shear stress and immune surveillance (18). Other important features of the subpopulations of aggressive CTCs are key for survival and include the resistance to anoikis and cancer stem cell (CSC)-like properties (19,20). In order to maximize survival odds in the hostile vasculature, rare CTC clusters or CTCs closely interacting via mucins with activated platelets and macrophages to form hetero-aggregates or circulating tumor microemboli (CTM) to facilitate adherence to the endothelium and trans-endothelial migration (14,18,21,22). Mucins are giant glycoproteins frequently upregulated in various cancers including NPC (23,24). The aberrant glycosylation of mucins result in the expression of Sialyl Lewis (sLe) structures, which promotes CTC dissemination (25,26). A MUC4 knockout mice model in breast cancer was cited here as an example to illustrate how mucins facilitated metastasis via enhancing interaction between CTCs with blood cells to augment CTC survival during hematogenous dissemination (27). MUC16 promoting metastasis functions as a ligand of selectin for binding with selectin-expressing white blood cells (WBCs) in pancreatic tumor cells (28). More in-depth discussion on the evidence for how the mucin-mediated immune modulation play crucial roles in tumor progression and metastasis was summarized nicely in some reviews (15,18). Studies in NPC revealing the CTC dissemination mechanisms mediated by EMT, mucins, or CTM are scanty and should be an important area for future CTC research to pursue. After extravasation, a CTC remains dormant and in this state of partial EMT, an intermediate phenotype with the highest plasticity for successful colonization of the secondary site, until the pro-tumorigenic immune niche emerges.
Liquid biopsy serial monitoring of treatment response and disease relapse in patients with locally advanced disease or metastases
For years, we rely on the traditional biopsy as the gold standard to understand the molecular basis of carcinogenesis. One of the limitations, particularly in NPC, is the difficulty to obtain enough primary tissues for characterization. Detailed study is also hampered by its highly heterogeneous nature and the heavy lymphocyte infiltration rendering NPC with a low carcinoma content. Traditional biopsy studies in solid tumors are hampered by spatial (multiple regions within a tumor) and temporal intratumor heterogeneity (ITH) during cancer evolution; this may generate misleading information resulting in underestimating the subclonal heterogeneity and drug resistance (29-32). Liquid biopsies offer the opportunities for identification of non-invasive biomarkers and have advantages of isolation from circulation and various biological fluids for real-time monitoring and prediction of treatment response. In the future, the liquid biopsy molecular tests will have the potential to become less expensive, quick, and easily repeatable to assess the dynamics of tumor heterogeneity to guide treatment decisions. It allows continuous follow-up for surveillance of recurrence and monitoring of MRD after surgery.
Most advanced cancer patients die from distant metastasis. CTCs and disseminated tumor cells (DTCs) shedding into the bloodstream are believed to be the metastatic seeds (33). In blood, CTC and cfDNA are the two most common entities of liquid biopsies (20,34). The numbers of CTCs are estimated to be ~1–10 amongst billions of red blood cells (RBCs) and millions of WBCs in 10 mL blood (Figure 1). Only a small fraction (<0.01%) of CTCs originating from primary lesions or distant metastases survive the hostile circulation environment and shear stresses resulting to damaged or fragmented or dying CTCs due to apoptosis, anoikis, and immune attack. CTC transit time is short in the circulation with only 1–2.4 hours (35). Enrichment of the rare CTCs from the blood is an extremely challenging task relying on sophisticated cellular isolation platforms. Analysis of the rare CTCs allow DNA, RNA, and protein characterization to understand the biological mechanisms of metastasis. Study of CTCs may lead to the knowledge of drug resistance mechanisms, which would guide clinicians for timely and precise treatment decision. Earlier studies indicated the presence of CTCs had a negative prognostic significance in breast, colorectal and prostate cancers using the CellSearch platform, the only FDA-approved technology for CTC enumeration in the clinic (36-38). Extensive CTC research on metastatic castration-resistant prostate cancer (mCRPC) has established CTC counts as a strong independent prognostic factor and the presence of the androgen receptor splice variant 7 (AR-v7), as a predictive biomarker to guide individualized treatment (39-41). Another example for potential application of CTCs in guidance of clinical management is the serial longitudinal monitoring of CTC DNA copy number variation (CNV) along treatment in small cell lung cancer that revealed predictive signatures of treatment response for systematic therapy (42). The combined analysis of both CTC and cfDNA may provide complementary information for prediction and prognosis of treatment responses and holds great potential for real-time monitoring of disease status and personalized medicine (20,34).
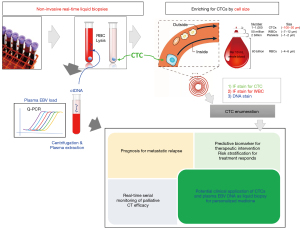
NPC CTC enrichment and enumeration
CTC isolation usually involves negative depletion of WBCs or positive enrichment strategies based on the biological properties i.e., using antibody-capture to recognize the tumor antigens expressed on the cell surface or physical properties including size-, density-, deformability-, or electric charge-based microfluidics approaches (34). Other technologies include high-throughput imaging to scan cells on slides such as the Epic and isolation based on functional characteristics such as the EPISPOT secretion assay (34). Previous CTC studies in better-studied breast, colon, and prostate cancers indicate CTC detection can be more sensitive, quantitative, and specific than conventional imaging or biomarker serology (43-45).
The technological advancement leads to many microfluidic devices for CTC enrichment, among which the underlying CTC enrichment technology based on size and hydrodynamic centrifugal forces provides the most gentle, fast and efficient method to isolate viable and label-free CTCs for multiple downstream applications, next-generation sequencing (NGS) mutational profiling, single-cell genomics and transcriptome analysis, and culturing for ex vivo drug testing in mouse xenografts (46,47). While label-free technologies are more reliable for CTC enrichment, they may be less specific compared to antigen-directed enrichment methods such as CellSearch or IsoFlux. Our group embarked on a pilot study and established the size-based microfluidic CTC enrichment platform for CTC enumeration and NGS analysis in eight cancers of importance to Hong Kong Chinese including breast, prostate, liver, lung, esophageal, gastric, colorectal and NPC (46,48-50). The larger size label-free viable CTCs travel to the outside of the microchannels of the spiral microfluidic devices using hydrodynamic forces and are, thus, rapidly depleted of the smaller WBCs in less than 1 hour for 7.5 mL blood (46). Our earlier work established a highly adaptable workflow after CTC enrichment to facilitate systematic CTC enumeration combining an immunofluorescent staining strategy and an automated CTC analytical pipeline with open-source image analyzer (50). CTC enumeration was with various cell surface markers including pan-cytokeratins (pan-CK), mesenchymal and stem cell markers independent of epithelial cell adhesion molecule (EpCAM) expression to identify CTCs. Compared to the CellSearch System, which is the only medical CTC isolation device approved by the US Food and Drug Administration (FDA), this allows the isolation of both epithelial and mesenchymal subpopulations of CTCs with CSC states that undergo partial EMT with loss or reduced epithelial markers such as EpCAM or pan-CK (19).
We performed literature search with the keywords “nasopharyngeal” and “CTC” in the National institute of Health (NIH) National Center for Biotechnology Information (NCBI) PubMed database (https://pubmed.ncbi.nlm.nih.gov/). With the aim to review the clinical utility of non-invasive real-time monitoring of CTC analysis for NPC treatment, ten recent publications focusing on CTC enumeration and mutational profiling were selected after manual inspection of abstracts. Table 1 summarizes CTC analysis in these ten recent NPC studies utilizing various methods including the well-known EpCAM-based CellSearch platform for positive selection (53-55) and others more frequently involving the negative enrichment strategies such as CD45 immuno-magnetic beads to deplete the WBCs (51,56), sized-based filtration [isolation by size of epithelial tumor cells (ISET), microsieve, CanPatrol] (52,57,58,60) and microfluidics (Clear Cell FX system) in our group (48) for CTC enrichment. Although these studies employed different CTC enrichment strategies, the majority of them (9/10) support the potential clinical utility of serial analysis of CTCs for real-time monitoring and prediction of treatment response for mNPC, when used together with imaging. Most studies showed that CTC counts were higher in mNPC compared to non-mNPC patients and in patients with poor prognosis (51-56). CTC counts is an independent prognostic biomarker with poor prognosis with no significant association with clinical parameters such as T and N stage (52,54,57). Seven out of these ten studies aimed to assess the predictive biomarker value for treatment response with recruitment schedule including serial blood sample collection before treatment, during and at post-treatment timepoints. Results suggested the post-treatment CTC findings or the favorable change of CTC status from positive baseline to negative by the end of treatment or a decrease of CTC counts were useful predictive markers for positive treatment response (48,52,55-57). Contrary to the other studies, an early study by Vo et al., used microsieve CTC enrichment in 50 NPC cases and concluded that EBV cfDNA outperformed CTC enumeration for prognostication (58). The two largest NPC CTC studies utilizing CellSearch for CTC enrichment and CK/EpCAM for enumeration of CTCs in 270 and 370 NPC cases reported the combined CTC and EBV cfDNA could be better predictive biomarkers based on the observation that CTCs could provide additional prognostic information by improvement of the specificity despite the inferior sensitivity compared to EBV cfDNA (54,55).
Table 1
| Reference | No. of NPC patients | Recruitment timeline | Enrichment methods | DNA/RNA/protein assays of CTC | CTC and plasma EBV DNA status | Comparison of CTC and EBV DNA | Real-time monitoring of disease progression and treatment outcome | Survival endpoint |
|---|---|---|---|---|---|---|---|---|
| Ko et al., 2020 (48) | 21 mNPC | Four timepoints at baseline, pre-II, pre-III, end of palliative CT | Size-based microfluidics | NGS: BML IF: EpCAM+/CK+/CD45− | Post-treatment BML not correlated with PFS positive CTC counts and plasma EBV DNA detected by Q-PCR analysis at baseline, post-treatment, and longitudinal change of CTCs associated with PFS | Yes | Yes | PFS |
| Liu et al., 2020 (51) | 178 locally advanced HNSCC (NPC + hypopharyngeal squamous cell carcinoma) | Pre- and post-treatment | Negative immunomagnetic bead enrichment | FISH | increased CTCs and CTC ≥2 | – | Yes | PFS |
| Yu et al., 2020 (52) | 179 NPC patients | Pre-treatment 75; post-treatment 179 | CanPatrol (filtration) | mRNA ISH: EpCAM/CK 8/18/19, vimentin, twist, CD45 FN1 | CTC >9, >5 mesenchymal CTCs, high FN1+ CTCs | – | Yes | PFS |
| Sun et al., 2020 (53) | 250 NPC cases, 136 mNPC and 114 non-mNPC | Pre-treatment | CellSearch | IF: EpCAM+/CK+/CD45− | Higher CTC counts in mNPC | – | – | – |
| Ou et al., 2019 (54) | 370 NPC cases; 288 stage IV and 70 stage III | Pre-treatment | CellSearch | IF: EpCAM+/CK+/CD45− | Higher CTC counts in stage IV versus III NPC; independent prognostic factor, combined CTC and EBV DNA detected by Q-PCR analysis better predict prognosis | Yes | – | – |
| You et al., 2019 (55) | 270 NPC; 122 non-mNPC, 148 mNPC | Pre- and post-treatment | CellSearch | IF: EpCAM+/CK+/CD45− | CTC counts and EBV DNA detected by Q-PCR are strong predictive biomarkers for mNPC | Yes | Yes | PFS and OS |
| Zhang et al., 2018 (56) | 50 NPC (29 M0, 21 relapse or M1) | Pre- and post-treatment | CD45 magnetic bead depletion of WBC | IF: EpCAM+/CD45− or CEP8 aneuploidy/CD45− subtraction enrichment immunostaining-fluorescence iFISH, karyotyping | CEP8 aneuploidy predict treatment efficacy | – | Yes | Relapse or distant metastasis |
| Li et al., 2018 (57) | 131 NPC M0 (39 stage I–II, 92 stage III–IV) | Pre- and 1-week post-treatment | CanPatrol (filtration) | RNA ISH: COX2, CK8/18/19 (epithelial CTC) vimentin/twist1 (mesenchymal CTC) | Post-treatment COX2+ CTC independent prognostic markers | – | Yes | PFS and OS |
| Vo et al., 2016 (58) | 46 NPC | Pre- and 1-month post-treatment | Microsieve (sized-based) | IF: CK+/CD45− (canonical CTC) CK−/CD45− (potential CTC) | CTC no association with clinical stage, EBV cfDNA by Q-PCR analysis outperforms CTC enumeration in prognosis of treatment outcome and OS | Yes | Yes | Treatment outcome/OS |
| He et al., 2017 (59) | 33 NPC | Pre-treatment | ISET (sized-based) | IF: CK5/6, P63 ISH: EBERs | CTC or CTM are detected in all stages of NPC. CTC correlated with EBV VCA-IgA and EBV DNA load | Yes | – | – |
CTCs, circulating tumor cells; EBV, Epstein-Barr virus; mNPC, metastatic nasopharyngeal carcinoma; HNSCC, head and neck squamous cell carcinoma; NPC, nasopharyngeal carcinoma; WBC, white blood cell; ISET, isolation by size of epithelial tumor cells; NGS, next-generation sequencing; BML, blood mutation load; EpCAM, epithelial cell adhesion molecule; CK, cytokeratins; FISH, fluorescence in situ hybridization; ISH, in situ hybridization; FN1, fibronectin 1; iFISH, interphase FISH; COX2, cyclooxygenase-2; EBERs, Epstein-Barr encoding regions; PFS, progression-free survival; Q-PCR, quantitative-polymerase chain reaction; OS, overall survival; VCA, viral capsid antigen; IgA, immunoglobulin A.
Canonical epithelial versus mesenchymal CTCs and CTM in NPC
Epithelial cells undergo EMT to gain motility, increase interaction with extracellular environment, and loss of apical-basal polarity and cell-cell contact (17,61). During EMT, the expression of an epithelial marker gene such as E-cadherin is reduced, while those of mesenchymal marker genes including fibronectin, N-cadherin, vimentin etc. are increased. EMT has been demonstrated to play a role in the serial CTC monitoring of breast cancer patients during CT and disease progression (62). Comparing the pre- and post-treatment blood samples, the increase of proportions of mesenchymal CTCs accompanied the treatment relapse and half of the patients with CTC counts decreased and switched to the epithelial phenotype in their responding tumors. The current understanding in the CTC field is that CTCs are highly heterogenous and remain in a partial EMT cell state in between epithelial and mesenchymal phenotypes, maintaining the highest plasticity to form distant metastasis (63). Only a subpopulation of aggressive CTCs with CSC-like properties are able to escape immune surveillance and undergo clonal evolution under the drug treatment selective pressure. Another key property for initiating of metastasis is CTC clusters, although little is known about their origin, underlying mechanisms to enter circulation and the multiple steps of the metastatic cascade (64). Dissemination of CTC clusters may be enhanced by the leaky blood vessels, through invadopodia or macrophage-mediated intravasation. The CTC clusters may be formed by two cells or multiple CTCs (<50–100 cells) with cellular heterogeneity or cooperation with stromal cells or platelets to provide a survival advantage against anoikis, shielding from shear forces, immune surveillance, and other environmental stresses (65-67). The detection of CTC clusters at baseline of treatment is associated with poor prognosis in various cancers including lung, melanoma, breast, colorectal, gastric, pancreatic, and head and neck cancers (64,68-74). It is not clear if the above-mentioned connection of CTC/EMT/CSC status plays a role in NPC CTC dissemination. NPC studies examining the EMT/CSC issue and CTM is scanty. In the literature, only one NPC study detected CTM by ISET (60) and two reports enriched CTCs by the filtration method with the CanPatrol platform or by depletion of WBCs with CD45 magnetic beads and utilization of RNA-in situ hybridization (ISH) techniques aimed to provide evidence that EMT is also involved in NPC CTC dissemination (52,57). Fibronectin is a stromal extracellular matrix (ECM) glycoprotein transmitting ECM signals via integrin. It is an EMT marker able to stimulate an EMT response (75,76). Application of CanPatrol CTC enrichment to capture both epithelial (EpCAM, CK) and mesenchymal (FN1) CTCs by filtration with an 8 µm membrane and RNA-ISH in 179 NPC patients, the authors revealed that more than two phenotypic mesenchymal CTCs with high-expression of FN1 and therapy (CT, radiotherapy, and CRT) remained as independent prognostic indicators of poor progression-free survival (PFS) in a multivariate Cox regression analysis (52). Another NPC study also utilizing the same CanPatrol platform and RNA-ISH methods for detection of CTC with cyclooxygenase-2 (COX2) overexpression (57). Li et al. examined the pre- and post-treatment CTC analyses that revealed that only the post-treatment CTC with COX2 expression was an independent prognostic indicator of PFS and OS (n=131) (57). The link of CTCs with COX2 is novel and was not previously observed in other cancer types. Further validation studies confined to patients receiving homogenous therapy are required to reveal the unclear independent prognostic role of mesenchymal CTCs with high expression of FN1 or CTC with COX2 expression as heterogenous therapies was a confounding factor, which could contribute to biased results. More studies are needed to improve the understanding of the mechanistic link between CTC/EMT/CSC and CTC clusters to identify novel therapeutics targets for precision medicine.
Potential complementary role of cfDNA and EBV plasma DNA in mNPC
Compared to other solid cancers, NPC is unique given the ubiquitous association and intimate role of latent EBV infection within the multi-step NPC tumorigenesis (77). The plasma cell-free EBV DNA (78-82) and microRNA (BARTs) (83-85), are utilized for NPC screening, prognostication, disease monitoring and surveillance of recurrence. The rationale of tracking tumor burden is based on the fact that NPC cells release shorter EBV DNA fragments (<181 bp) during apoptosis compared to those released during viral lytic phase (86,87). Various EBV genome targets include the most commonly used multiple copy target (BamHI-W) and a single-copy EBV gene, Epstein-Barr virus nuclear antigen 1 (EBNA-1). Since the early reports of the detection of serum or plasma EBV DNA in NPC patients (78,79), numerous studies validated and refined its usefulness for clinical applications, guidance for selection of locally advanced patients for adjuvant CT after concurrent chemoradiation, prognostication, staging, surveillance and screening (80-82,88,89). Several excellent overviews detailly review and discuss the current challenges and enormous potential clinical application of plasma EBV DNA for prognostication, risk stratification and post-treatment surveillance (87,90-92).
The survival of mNPC patients diagnosed with advanced refractory or metastatic disease is dismal. Obtaining conventional invasive biopsies at multiple timepoints and lesions for disease monitoring is not practically feasible. NPC studies is also hampered by the inherently small and highly heterogenous nature of biopsies. To date, no study has reported CTC molecular profiling. In our pilot NPC CTC studies, we collected 7.5 mL blood from patients and isolated the CTCs based on size separation with the FX microfluidic chips. NPC CTC mutational profiling was deep-sequenced after whole-genome amplification, standard library preparation and hybridization-based capture of an oncology panel of 1,366 genes with the 7M NimbleGen capture kits. Serial longitudinal examination at four timepoints (baseline, pre-II, pre-III, and post-treatment) to reveal the dynamics and the molecular features of 21 mNPC patients treated by palliative CT with 11 synchronous and 10 metachronous metastasis (48). Radiologic imaging showed that 9 patients had poor response and 12 patients had good response (4 with partial and 8 with complete response). The FX CTC isolation provided an average recovery rate of 70% in cell line spike-in experiments. The CTC SeqCap NGS mutation profiling protocol and bioinformatics pipeline was validated with a cell line with known mutations by spike-in experiments. Somatic mutations were detected with high precision with a minimum purity of 2.5%. Positive blood mutation load (BML) was set at >0.57 mutations per Mb and was detected in 13 NPC patients. The patients with positive post-treatment BML associated with a trend of shorter PFS (P=0.174). Positive CTC status frequency was 55% at baseline, 50% at pre-II, 61% at pre-III and 67% at post-treatment. Positive EBV status frequency was 81% at baseline (cut-off of 1,500 copies/mL), 65% at pre-II, 58% at pre-III, and 55% at post-treatment (cut-off 60 copies/mL for during and post-treatment timepoints). Positive plasma EBV status at baseline, during and post-treatment, as well as positive baseline and post-treatment CTC status, were associated significantly with shorter PFS. The longitudinal changes in CTCs and EBV DNA along treatment was predictive for disease relapse. Liquid biopsy longitudinal real-time monitoring with EBV DNA, CTC counts or BML were concordant with imaging for the 9 mNPC patients with poor response. CTC molecular analysis was an alternative means for tracking tumor burden in 75% (3/4) mNPC patients with partial response with negative or ambiguous EBV findings during and at post-treatment. Four out of eight patients with complete responses assessed by post-treatment PET-CT imaging recurred after 8–24 months. The liquid biopsy real-time monitoring findings (both EBV DNA by Q-PCR analysis and CTC enumeration and mutation burden analysis) of these mNPC patients provide complementary evidence as a more sensitive means of tracking MRD earlier than conventional imaging. The potential clinical application of combining the CTC and EBV DNA in mNPC is summarized in Figure 1. The refractory locally advanced and mNPC patients are treated by systemic therapies and the treatment response is assessed by imaging, which is not adequate to track changes in tumor burden. We have demonstrated the combined molecular analysis of liquid biopsy i.e., CTC enumeration and plasma EBV load in a pilot study of 21 mNPC patients receiving palliative CT provided additional prognostic information and earlier detection of MRD used in conjunction with imaging (48). In addition to mNPC, we have shown that the liquid biopsy serial monitoring of treatment response and disease relapse by combining the changes of baseline and pre-cycle III CTC counts and cfDNA level may has useful clinical utility for prediction and prognosis of CT response in advanced esophageal squamous cell carcinoma (ESCC) patients (49). CTC counts and EBV load are both sources of liquid biopsy diagnostics having different but complementary information for prognostication and prediction of treatment response, should be utilized together with imaging to improve the accuracy for personalized medicine and guide treatment decision (20,34,93).
Among the ten NPC CTC studies, three reports (53,54,59) only analyzed CTCs at a single timepoint before treatment, while the other seven studies performed serial monitoring in NPC (Table 1). Our study is the only one that performed serial monitoring during palliative treatment at four timepoints, including baseline, pre-cycle II, pre-cycle III, and end of treatment, while six studies collected blood samples at two timepoints pre- and post-treatment (49,51-58). CTC enrichment was based on surface-marker-based capture (CellSearch platform) (53-55); and marker independent platforms including CanPatrol (52,57), microfluidics (48), Microsieve (58), and ISET (59), and WBC depletion strategy (51,56). Half of these studies analyzed both CTC enumeration and EBV DNA by quantitative PCR to evaluate their clinical utility alone or in combination (48,54,55,58,60). Among these, three real-time monitoring NPC studies by Ou et al. (n=370 stage III and IV NPC), Ko et al. (n=21 mNPC), and You et al. (n=270 non-mNPC and mNPC), suggested the potential complementary prognostic and predictive roles of combined CTC and EBV DNA (48,54,55), while Vo et al. (n=46 NPC) concluded EBV DNA outperformed enumeration in prognostication (58). He et al. included 33 NPC patients and detected the presence of CTCs in all stages, but no association with the TNM stages was observed (59). They reported the observation of CTM in addition to CTC enumeration, which showed positive correlation with EBV activation by EBV viral capsid antigen (VCA)-immunoglobulin A (IgA) and EBV DNA levels. To date, You et al. performed the largest CTC NPC study with 270 NPC cases for real-time monitoring of treatment responses and provided evidence that NPC patients with conversion of unfavorable CTC and EBV DNA from baseline CTC enumeration (cut-off =9, EBV =10,000 copies) to favorable post-treatment CTC counts (cuf-off =1 and EBV =4,000 copies) had a longer PFS and OS compared to those with unfavorable CTCs and EBV DNA status at baseline and post-treatment (55). For mNPC patients with good response assessed by imaging, those with favorable post-treatment CTCs and EBV DNA had significantly longer PFS and OS. The largest sample size study by Ou et al. (n=370) enriched CTCs by the CellSearch platform and detected higher CTC counts in NPC patients with stage IV versus III with an estimated cut-off of 0.5; CTC numbers were an independent prognostic factor (54). The observation of higher CTC counts in mNPC was substantiated by Sun et al. (n=250), who performed CTC enrichment also by the CellSearch platform (53). Liu et al. enriched CTCs by CD45 depletion methods and CTCs were enumerated by mRNA fluorescence in situ hybridization (FISH) (51). Zhang et al. also enriched CTCs by the CD45 depletion method and CTCs enumerated by the DNA level CEP8 aneuploidy were predictive of treatment efficacy (56). Yu et al. enriched CTCs by CanPatrol based on filtration strategy and demonstrated the presence of mesenchymal CTCs by mRNA ISH with fibronectin 1, vimentin and twist, in addition to epithelial EpCAM/CK transcripts (52). Using a similar CTC enrichment and enumeration approach, Li et al. reported the COX2-positive CTC number was an independent prognostic marker in a cohort of 131 NPC patients (57). The three largest NPC CTC studies (53-55) utilized the CellSearch platform for CTC isolation, which depends on EpCAM expression for CTC capture. With the aim to improve the sensitivity of CTC assays, and to further study CTCs which are highly heterogenous, future studies with CTC isolation strategies independent of cell-surface markers are warranted. More studies are also required to explore the utilization of a cocktail of mRNA transcripts of epithelial and mesenchymal markers, as well as EBV genomic fragments such as BamHI-W as probes for FISH or EBNA-1 protein expression for the identification of CTCs and proof of its NPC tumor origins.
Conclusions
Longitudinal liquid biopsy serial monitoring of combined CTC counts and EBV DNA may provide additional information that may prove complementary for that provided by CTCs and may precede imaging results by weeks or even months for detection of minimal disease burden and prognostication in mNPC patients with advanced disease, when accompanied with imaging. Although the NPC CTC field is still in its infancy, with development of sophisticated CTC isolation platforms and sensitive multi-omics profiling technological advancement, the potential clinical application of CTC analysis on its own or in combination with other sources of liquid biopsy holds great promise in future translation into the clinic, as non-invasive real-time monitoring of mNPC treatment for precision personalized medicine.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Maria Li. Lung and Lawrence S. Young) for the series “NPC Biomarkers” published in Annals of Nasopharynx Cancer. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://anpc.amegroups.com/article/view/10.21037/anpc-21-7/coif). The series “NPC Biomarkers” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma. Lancet 2016;387:1012-24. [Crossref] [PubMed]
- Hong Kong Cancer Registry. Overview of Hong Kong Cancer Statistics of 2018. Hong Kong Hospital Authority; 2021. (accessed on 17 May 2021). Available online: https://www3.ha.org.hk/cancereg/
- Chinese Journal of Cancer. The 150 most important questions in cancer research and clinical oncology series: questions 6-14: Edited by Chinese Journal of Cancer. Chin J Cancer 2017;36:33.
- Lee AW, Poon YF, Foo W, et al. Retrospective analysis of 5037 patients with nasopharyngeal carcinoma treated during 1976-1985: overall survival and patterns of failure. Int J Radiat Oncol Biol Phys 1992;23:261-70. [Crossref] [PubMed]
- Li AC, Xiao WW, Shen GZ, et al. Distant metastasis risk and patterns of nasopharyngeal carcinoma in the era of IMRT: long-term results and benefits of chemotherapy. Oncotarget 2015;6:24511-21. [Crossref] [PubMed]
- Loong HH, Ma BB, Chan AT. Update on the management and therapeutic monitoring of advanced nasopharyngeal cancer. Hematol Oncol Clin North Am 2008;22:1267-78. x. [Crossref] [PubMed]
- Li YY, Chung GT, Lui VW, et al. Exome and genome sequencing of nasopharynx cancer identifies NF-κB pathway activating mutations. Nat Commun 2017;8:14121. [Crossref] [PubMed]
- Zheng H, Dai W, Cheung AK, et al. Whole-exome sequencing identifies multiple loss-of-function mutations of NF-κB pathway regulators in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A 2016;113:11283-8. [Crossref] [PubMed]
- Deng M, Dai W, Yu VZ, et al. Cylindromatosis Lysine 63 Deubiquitinase (CYLD) Regulates NF-kB Signaling Pathway and Modulates Fibroblast and Endothelial Cells Recruitment in Nasopharyngeal Carcinoma. Cancers (Basel) 2020;12:1924. [Crossref] [PubMed]
- Cheung AK, Ko JM, Lung HL, et al. Cysteine-rich intestinal protein 2 (CRIP2) acts as a repressor of NF-kappaB-mediated proangiogenic cytokine transcription to suppress tumorigenesis and angiogenesis. Proc Natl Acad Sci U S A 2011;108:8390-5. [Crossref] [PubMed]
- Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet 2014;46:866-71. [Crossref] [PubMed]
- Miao Z, Ali A, Hu L, et al. Microtubule actin cross-linking factor 1, a novel potential target in cancer. Cancer Sci 2017;108:1953-8. [Crossref] [PubMed]
- Chan KCA, Woo JKS, King A, et al. Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. N Engl J Med 2017;377:513-22. [Crossref] [PubMed]
- Smith HA, Kang Y. The metastasis-promoting roles of tumor-associated immune cells. J Mol Med (Berl) 2013;91:411-29. [Crossref] [PubMed]
- Marimuthu S, Rauth S, Ganguly K, et al. Mucins reprogram stemness, metabolism and promote chemoresistance during cancer progression. Cancer Metastasis Rev 2021;40:575-88. [Crossref] [PubMed]
- Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell 2013;23:272-3. [Crossref] [PubMed]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2:442-54. [Crossref] [PubMed]
- Ganguly K, Rauth S, Marimuthu S, et al. Unraveling mucin domains in cancer and metastasis: when protectors become predators. Cancer Metastasis Rev 2020;39:647-59. [Crossref] [PubMed]
- Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611-29. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov 2016;6:479-91. [Crossref] [PubMed]
- Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158:1110-22. [Crossref] [PubMed]
- Dive C, Brady G. SnapShot: Circulating Tumor Cells. Cell 2017;168:742-742.e1. [Crossref] [PubMed]
- Kuo WR, Tsai SM, Jong SB, et al. Significance of tumour markers in nasopharyngeal carcinoma. J Otolaryngol 1996;25:32-6. [PubMed]
- Fernandes E, Sores J, Cotton S, et al. Esophageal, gastric and colorectal cancers: Looking beyond classical serological biomarkers towards glycoproteomics-assisted precision oncology. Theranostics 2020;10:4903-28. [Crossref] [PubMed]
- Schultz MJ, Swindall AF, Bellis SL. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev 2012;31:501-18. [Crossref] [PubMed]
- Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015;15:540-55. [Crossref] [PubMed]
- Rowson-Hodel AR, Wald JH, Hatakeyama J, et al. Membrane Mucin Muc4 promotes blood cell association with tumor cells and mediates efficient metastasis in a mouse model of breast cancer. Oncogene 2018;37:197-207. [Crossref] [PubMed]
- Chen SH, Dallas MR, Balzer EM, et al. Mucin 16 is a functional selectin ligand on pancreatic cancer cells. FASEB J 2012;26:1349-59. [Crossref] [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. [Crossref] [PubMed]
- Hao JJ, Lin DC, Dinh HQ, et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet 2016;48:1500-7. [Crossref] [PubMed]
- de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. [Crossref] [PubMed]
- Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256-9. [Crossref] [PubMed]
- Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell 2013;23:573-81. [Crossref] [PubMed]
- Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4:650-61. [Crossref] [PubMed]
- Meng S, Tripathy D, Frenkel EP, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res 2004;10:8152-62. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Cohen SJ, Alpaugh RK, Gross S, et al. Isolation and characterization of circulating tumor cells in patients with metastatic colorectal cancer. Clin Colorectal Cancer 2006;6:125-32. [Crossref] [PubMed]
- Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res 2007;13:7053-8. [Crossref] [PubMed]
- Scher HI, Lu D, Schreiber NA, et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol 2016;2:1441-9. [Crossref] [PubMed]
- Scher HI, Graf RP, Schreiber NA, et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol 2018;4:1179-86. [Crossref] [PubMed]
- Lorente D, Olmos D, Mateo J, et al. Circulating tumour cell increase as a biomarker of disease progression in metastatic castration-resistant prostate cancer patients with low baseline CTC counts. Ann Oncol 2018;29:1554-60. [Crossref] [PubMed]
- Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 2017;23:114-9. [Crossref] [PubMed]
- Budd GT, Cristofanilli M, Ellis MJ, et al. Circulating tumor cells versus imaging--predicting overall survival in metastatic breast cancer. Clin Cancer Res 2006;12:6403-9. [Crossref] [PubMed]
- Pantel K, Alix-Panabières C. The potential of circulating tumor cells as a liquid biopsy to guide therapy in prostate cancer. Cancer Discov 2012;2:974-5. [Crossref] [PubMed]
- Hardingham JE, Grover P, Winter M, et al. Detection and Clinical Significance of Circulating Tumor Cells in Colorectal Cancer--20 Years of Progress. Mol Med 2015;21:S25-31. [Crossref] [PubMed]
- Warkiani ME, Khoo BL, Wu L, et al. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat Protoc 2016;11:134-48. [Crossref] [PubMed]
- Khoo BL, Warkiani ME, Tan DS, et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PLoS One 2014;9:e99409. [Crossref] [PubMed]
- Ko JM, Vardhanabhuti V, Ng WT, et al. Clinical utility of serial analysis of circulating tumour cells for detection of minimal residual disease of metastatic nasopharyngeal carcinoma. Br J Cancer 2020;123:114-25. [Crossref] [PubMed]
- Ko JMY, Ng HY, Lam KO, et al. Liquid Biopsy Serial Monitoring of Treatment Responses and Relapse in Advanced Esophageal Squamous Cell Carcinoma. Cancers (Basel) 2020;12:1352. [Crossref] [PubMed]
- Wong VC, Ko JM, Lam CT, et al. Succinct workflows for circulating tumor cells after enrichment: From systematic counting to mutational profiling. PLoS One 2017;12:e0177276. [Crossref] [PubMed]
- Liu K, Chen N, Wei J, et al. Clinical significance of circulating tumor cells in patients with locally advanced head and neck squamous cell carcinoma. Oncol Rep 2020;43:1525-35. [Crossref] [PubMed]
- Yu Y, Lin ZX, Li HW, et al. Circulating Tumor Cells and Fibronectin 1 in the Prognosis of Nasopharyngeal Carcinoma. Technol Cancer Res Treat 2020;19:1533033820909911. [Crossref] [PubMed]
- Sun L, Wang Y, Shi J, et al. Association of Plasma Epstein-Barr Virus LMP1 and EBER1 with Circulating Tumor Cells and the Metastasis of Nasopharyngeal Carcinoma. Pathol Oncol Res 2020;26:1893-901. [Crossref] [PubMed]
- Ou G, Xing S, Li J, et al. Circulating tumor cells: a valuable marker of poor prognosis for advanced nasopharyngeal carcinoma. Mol Med 2019;25:50. [Crossref] [PubMed]
- You R, Liu YP, Lin M, et al. Relationship of circulating tumor cells and Epstein-Barr virus DNA to progression-free survival and overall survival in metastatic nasopharyngeal carcinoma patients. Int J Cancer 2019;145:2873-83. [Crossref] [PubMed]
- Zhang J, Shi H, Jiang T, et al. Circulating tumor cells with karyotyping as a novel biomarker for diagnosis and treatment of nasopharyngeal carcinoma. BMC Cancer 2018;18:1133. [Crossref] [PubMed]
- Li YJ, Luo Y, Xie XQ, et al. The prognostic value of COX-2 expression on circulating tumor cells in nasopharyngeal carcinoma: A prospective analysis. Radiother Oncol 2018;129:396-402. [Crossref] [PubMed]
- Vo JH, Nei WL, Hu M, et al. Comparison of Circulating Tumour Cells and Circulating Cell-Free Epstein-Barr Virus DNA in Patients with Nasopharyngeal Carcinoma Undergoing Radiotherapy. Sci Rep 2016;6:13. [Crossref] [PubMed]
- He C, Huang X, Su X, et al. The association between circulating tumor cells and Epstein-Barr virus activation in patients with nasopharyngeal carcinoma. Cancer Biol Ther 2017;18:888-94. [Crossref] [PubMed]
- Siraj AK, Masoodi T, Bu R, et al. MED12 is recurrently mutated in Middle Eastern colorectal cancer. Gut 2018;67:663-71. [PubMed]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. [Crossref] [PubMed]
- Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580-4. [Crossref] [PubMed]
- Agnoletto C, Corrà F, Minotti L, et al. Heterogeneity in Circulating Tumor Cells: The Relevance of the Stem-Cell Subset. Cancers (Basel) 2019;11:483. [Crossref] [PubMed]
- Giuliano M, Shaikh A, Lo HC, et al. Perspective on Circulating Tumor Cell Clusters: Why It Takes a Village to Metastasize. Cancer Res 2018;78:845-52. [Crossref] [PubMed]
- Cho EH, Wendel M, Luttgen M, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol 2012;9:016001. [Crossref] [PubMed]
- Duda DG, Duyverman AM, Kohno M, et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proc Natl Acad Sci U S A 2010;107:21677-82. [Crossref] [PubMed]
- Stott SL, Lee RJ, Nagrath S, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med 2010;2:25ra23. [Crossref] [PubMed]
- Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. [Crossref] [PubMed]
- Long E, Ilie M, Bence C, et al. High expression of TRF2, SOX10, and CD10 in circulating tumor microemboli detected in metastatic melanoma patients. A potential impact for the assessment of disease aggressiveness. Cancer Med 2016;5:1022-30. [Crossref] [PubMed]
- Mu Z, Wang C, Ye Z, et al. Prospective assessment of the prognostic value of circulating tumor cells and their clusters in patients with advanced-stage breast cancer. Breast Cancer Res Treat 2015;154:563-71. [Crossref] [PubMed]
- Zhang D, Zhao L, Zhou P, et al. Circulating tumor microemboli (CTM) and vimentin+ circulating tumor cells (CTCs) detected by a size-based platform predict worse prognosis in advanced colorectal cancer patients during chemotherapy. Cancer Cell Int 2017;17:6. [Crossref] [PubMed]
- Zheng X, Fan L, Zhou P, et al. Detection of Circulating Tumor Cells and Circulating Tumor Microemboli in Gastric Cancer. Transl Oncol 2017;10:431-41. [Crossref] [PubMed]
- Chang MC, Chang YT, Chen JY, et al. Clinical Significance of Circulating Tumor Microemboli as a Prognostic Marker in Patients with Pancreatic Ductal Adenocarcinoma. Clin Chem 2016;62:505-13. [Crossref] [PubMed]
- Fanelli MF, Oliveira TB, Braun AC, et al. Evaluation of incidence, significance, and prognostic role of circulating tumor microemboli and transforming growth factor-β receptor I in head and neck cancer. Head Neck 2017;39:2283-92. [Crossref] [PubMed]
- Lee JM, Dedhar S, Kalluri R, et al. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 2006;172:973-81. [Crossref] [PubMed]
- Park J, Schwarzbauer JE. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene 2014;33:1649-57. [Crossref] [PubMed]
- Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell 2004;5:423-8. [Crossref] [PubMed]
- Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 1999;59:1188-91. [PubMed]
- Mutirangura A, Pornthanakasem W, Theamboonlers A, et al. Epstein-Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res 1998;4:665-9. [PubMed]
- Chan KC, Hung EC, Woo JK, et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer 2013;119:1838-44. [Crossref] [PubMed]
- Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. N Engl J Med 2018;378:973. [Crossref] [PubMed]
- Qu H, Huang Y, Zhao S, et al. Prognostic value of Epstein-Barr virus DNA level for nasopharyngeal carcinoma: a meta-analysis of 8128 cases. Eur Arch Otorhinolaryngol 2020;277:9-18. [Crossref] [PubMed]
- Lu T, Guo Q, Lin K, et al. Circulating Epstein-Barr virus microRNAs BART7-3p and BART13-3p as novel biomarkers in nasopharyngeal carcinoma. Cancer Sci 2020;111:1711-23. [Crossref] [PubMed]
- Ramayanti O, Verkuijlen SAWM, Novianti P, et al. Vesicle-bound EBV-BART13-3p miRNA in circulation distinguishes nasopharyngeal from other head and neck cancer and asymptomatic EBV-infections. Int J Cancer 2019;144:2555-66. [Crossref] [PubMed]
- Zhang G, Zong J, Lin S, et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer 2015;136:E301-12. [Crossref] [PubMed]
- Chan KC, Zhang J, Chan AT, et al. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res 2003;63:2028-32. [PubMed]
- Lam WKJ, Chan KCA, Lo YMD. Plasma Epstein-Barr virus DNA as an archetypal circulating tumour DNA marker. J Pathol 2019;247:641-9. [Crossref] [PubMed]
- Leung SF, Chan KC, Ma BB, et al. Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann Oncol 2014;25:1204-8. [Crossref] [PubMed]
- Tang LQ, Chen QY, Fan W, et al. Prospective study of tailoring whole-body dual-modality 18Ffluorodeoxyglucose positron emission tomography/computed tomography with plasma Epstein-Barr virus DNA for detecting distant metastasis in endemic nasopharyngeal carcinoma at initial staging. J Clin Oncol 2013;31:2861-9. [Crossref] [PubMed]
- Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014;406:6499-512. [Crossref] [PubMed]
- Kim KY, Le QT, Yom SS, et al. Clinical Utility of Epstein-Barr Virus DNA Testing in the Treatment of Nasopharyngeal Carcinoma Patients. Int J Radiat Oncol Biol Phys 2017;98:996-1001. [Crossref] [PubMed]
- Tan R, Phua SKA, Soong YL, et al. Clinical utility of Epstein-Barr virus DNA and other liquid biopsy markers in nasopharyngeal carcinoma. Cancer Commun (Lond) 2020;40:564-85. [Crossref] [PubMed]
- Alix-Panabières C, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med 2012;63:199-215. [Crossref] [PubMed]
Cite this article as: Ko JMY. Clinical utility of circulating tumor cells for real-time serial monitoring of nasopharyngeal carcinoma patients. Ann Nasopharynx Cancer 2022;6:7.


